Research
We explore the principals of vertebrate development in the African clawed frog Xenopus, in which nuclear reprogramming of somatic cells had been originally pioneered (see www.xenbase.org ). Among the many advantages of Xenopus, we appreciate its ease of manipulation, the availibility of primary embryonic stem cells, and the unique possibility to combine embryological and reverse genetics approaches with biochemical analyses of virtually every developmental stage.
We investigate the hypothesis that changes in chromatin structure and composition define distinct states of cellular competence that constrain inductive processes and propel development in a directional manner. Over the years, this hypothesis has led us to seminal observations on specific developmental functions for H1 linker histones, ATP-dependent chromating remodelling machines, and histone modifying enzymes.
In recent years, a major focus of our work has become the analysis of stage-specific histone modification profiles, which impact embryonic stem cell behaviour and cell differentiation. These profiles visualize the histone modification landscape in embryos and reflect the maturation of chromatin, as cells pass from pluripotency to differentiated somatic cell states. We currently investigate a potential regulatory role for the cell cycle in chromatin maturation, which we believe to affect the writing and maintenance of histone modifications that regulate embryonic gene expression programs.
https://orcid.org/0000-0002-8068-9957
December 2020
H4K20 Methylation Is Differently Regulated by Dilution and Demethylation in Proliferating and Cell-Cycle-Arrested Xenopus Embryos
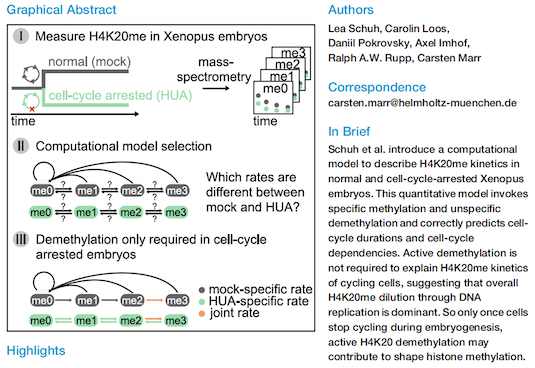
For more information see DOI: https://doi.org/10.1016/j.cels.2020.11.003
August 2018
The Xenopus animal cap transcriptome: building a mucociliary epithelium
With the availability of deep RNA sequencing, model organisms such as Xenopus offer an outstanding opportunity to investigate the genetic basis of vertebrate organ formation from its embryonic beginnings. Here we investigate dynamics of the RNA landscape during formation of the Xenopus tropicalis larval epidermis. Differentiation of non-neural ectoderm starts at gastrulation and takes about one day to produce a functional mucociliary epithelium, highly related to the one in human airways. To obtain RNA expression data, uncontaminated by non-epidermal tissues of the embryo, we use prospective ectodermal explants called Animal Caps (ACs), which differentiate autonomously into a ciliated epidermis. Their global transcriptome is investigated at three key timepoints, with a cumulative sequencing depth of ∼108 reads per developmental stage. This database is provided as online Web Tool to the scientific community. In this paper, we report on global changes in gene expression, an unanticipated diversity of mRNA splicing isoforms, expression patterns of repetitive DNA Elements, and the complexity of circular RNAs during this process. Computationally we derive transcription factor hubs from this data set, which may help in the future to define novel genetic drivers of epidermal differentiation in vertebrates.
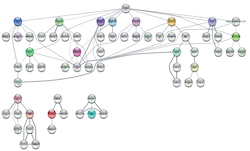
Transcription factor network potentially driving epidermal differentiation by Mogrify.
For more information see DOI: https://doi.org/10.1093/nar/gky771
Access to the database Xenopus Skin Differentiation
May 2017
Brg1 chromatin remodeling ATPase balances germ layer patterning by amplifying the transcriptional burst at midblastula transition
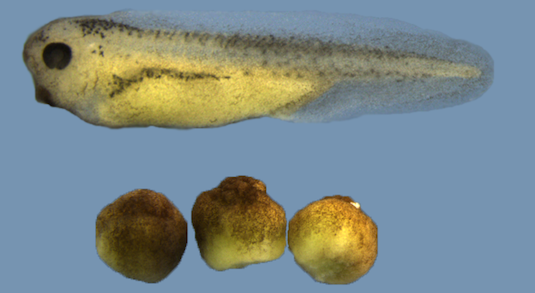
BRG1 depleted embryos fail to establish axial patterning. On top: wildtype embryo; on bottom: Brg1-depleted sibling embryos (after hatching).
Zygotic gene expression programs control cell differentiation in vertebrate development. In Xenopus, these programs are initiated by local induction of regulatory genes through maternal signaling activities in the wake of zygotic genome activation (ZGA) at the midblastula transition (MBT). These programs lay down the vertebrate body plan through gastrulation and neurulation, and are accompanied by massive changes in chromatin structure, which increasingly constrain cellular plasticity. Here we report on developmental functions for Brahma related gene 1 (Brg1), a key component of embyronic SWI/SNF chromatin remodeling complexes. Carefully controlled, global Brg1 protein depletion in X. tropicalis and X. laevis causes embryonic lethality or developmental arrest from gastrulation on. Transcriptome analysis at late blastula, before development becomes arrested, indicates predominantly a role for Brg1 in transcriptional activation of a limited set of genes involved in pattern specification processes and nervous system development. Mosaic analysis by targeted microinjection defines Brg1 as an essential amplifier of gene expression in dorsal (BCNE/Nieuwkoop Center) and ventral (BMP/Vent) signaling centers. Moreover, Brg1 is required and sufficient for initiating axial patterning in cooperation with maternal Wnt signaling. In search for a common denominator of Brg1 impact on development, we have quantitatively filtered global mRNA fluctuations at MBT. The results indicate that Brg1 is predominantly required for genes with the highest burst of transcriptional activity. Since this group contains many key developmental regulators, we propose Brg1 to be responsible for raising their expression above threshold levels in preparation for embryonic patterning.
For more information see DOI: https://doi.org/10.1371/journal.pgen.1006757
January 2013
Suv4-20h histone methyltransferases promote neuroectodermal differentiation by silencing the pluripotency-associated Oct-25 gene
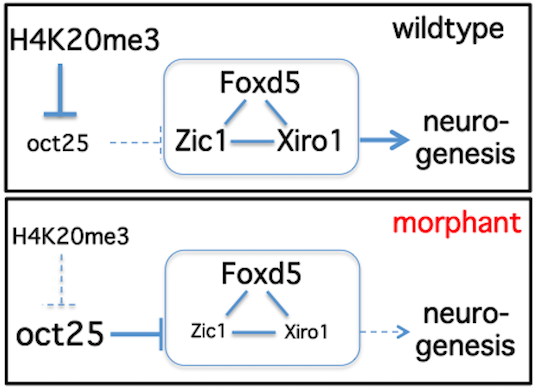
Model explaining the role of H4K20me3-mediated silencing of Oct-25 in the context of neural ground state formation
Post-translational modifications (PTMs) of histones exert fundamental roles in regulating gene expression. During development, groups of PTMs are constrained by unknown mechanisms into combinatorial patterns, which facilitate transitions from uncommitted embryonic cells into differentiated somatic cell lineages. Here we show that Xenopus laevis Suv4-20h1 and h2 histone methyltransferases (HMTases) are essential for induction and differentiation of the neuroectoderm. Interestingly, morphant embryos fail to repress the Oct4-related Xenopus gene Oct-25. We validate Oct-25 as a direct target of xSu4-20h enzyme mediated gene repression, showing by chromatin immunoprecipitaton that it is decorated with the H4K20me3 mark downstream of the promoter in normal, but not in double-morphant, embryos. Since knockdown of Oct-25 protein significantly rescues the neural differentiation defect in xSuv4-20h double-morphant embryos, we conclude that the epistatic relationship between Suv4-20h enzymes and Oct-25 controls the transit from pluripotent to differentiation-competent neural cells.
For more information see DOI: https://doi.org/10.1093/nar/gky771
July 2011
Stage-Specific Histone Modification Profiles Reveal Global Transitions in the Xenopus Embryonic Epigenome
The heatmap visualizes clusters of histone modifications according to their relative abundance at the four developmental stages, which we have investigated. The hierarchical clustering analysis produces a dendrogram, shown on the left side, with four major branches that correspond to specific developmental stages. Modifications associated with transcriptional activity are highlighted in green, while modifications associated with repressed/silent states are shown in magenta. The four clusters define histone modifications profiles (HMPs), which reflect the gradual transition from uncommitted to determined cell fates.Vertebrate embryos are derived from a transitory pool of pluripotent cells. By the process of embryonic induction, these precursor cells are assigned to specific fates and differentiation programs. Histone post-translatio-nal modifications are thought to play a key role in the establishment and maintenance of stable gene expression patterns underlying these processes. While on gene level histone modifications are known to change during differentiation, very little is known about the quantitative fluctuations in bulk histone modifications during development. To investigate this issue we analysed histones isolated from four different developmental stages of Xenopus laevis by mass spectrometry. In toto, we quantified 59 modification states on core histones H3 and H4 from blastula to tadpole stages. During this developmental period, we observed in general an increase in the unmodified states, and a shift from histone modifications associated with transcriptional activity to transcriptionally repressive histone marks. We also compared these naturally occurring patterns with 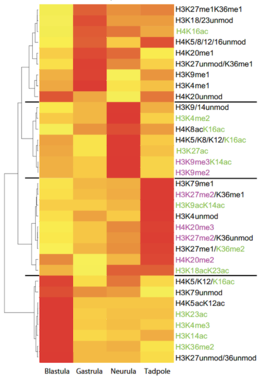 the histone modifications of murine ES cells, detecting large differences in the methylation patterns of histone H3 lysines 27 and 36 between pluripotent ES cells and pluripotent cells from Xenopus blastulae. By combining all detecte modification transitions we could cluster their patterns according to their embryonic origin, defining specific histone modification profiles (HMPs) for each developmental stage. To our knowledge, this data set represents the first compendium of covalent histone modifi-cations and their quantitative flux during normogenesis in a vertebrate model organism. The HMPs indicate a stepwise maturation of the embryonic epigenome, which may be causal to the progress-sing restriction of cellular potency during development.
the histone modifications of murine ES cells, detecting large differences in the methylation patterns of histone H3 lysines 27 and 36 between pluripotent ES cells and pluripotent cells from Xenopus blastulae. By combining all detecte modification transitions we could cluster their patterns according to their embryonic origin, defining specific histone modification profiles (HMPs) for each developmental stage. To our knowledge, this data set represents the first compendium of covalent histone modifi-cations and their quantitative flux during normogenesis in a vertebrate model organism. The HMPs indicate a stepwise maturation of the embryonic epigenome, which may be causal to the progress-sing restriction of cellular potency during development.
Stage-specific histone modification profiles.
For more information see DOI: https://doi.org/10.1371/journal.pone.0022548

