Kurat Group
Mechanisms of Chromatin Replication
The entire chromosome must be replicated and reassembled into daughter chromatids. Thus, the accurate duplication of eukaryotic chromosomes in the S phase of the cell cycle, consisting of genomic DNA and associated proteins, is one of the most fundamental processes of dividing cells. Errors in this process as well as changes in replication fork rates can be catastrophic and might result in “out-of-control” cells with the potential to cause diseases like cancer. It is largely unknown, how the eukaryotic replication machinery deals with chromatin during replication, how chromatin structure changes, how nucleosomes are disassembled in front, and reassembled in the back of the replicative helicase or to what extent chromatin factors and histone marks affect the progression of the replication fork through chromatin. Using purified proteins, I recently established a novel biochemical chromatin replication system with the potential to answer these fundamental questions directly (Kurat et al., 2017). My lab will use this in vitro system in combination with in vivo cell biological approaches, genomic and proteomic screens as well as structural analyses to determine
how chromatin replication rates are achieved on a global level and how this is regulated under normal conditions and under replication stress.
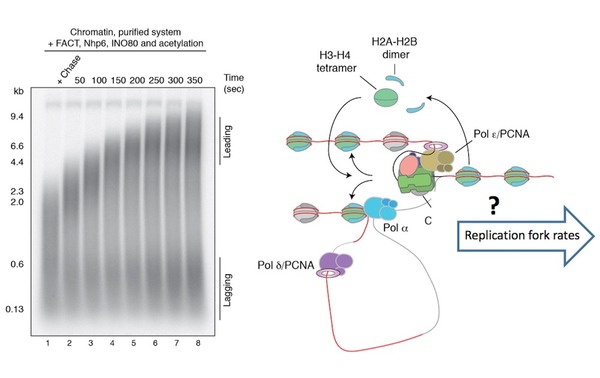
Figure modified from Kurat et al., Mol Cell. 2017 Jan 5;65(1):117-130. Left panel shows a chromatin replication reaction in vitro using ~ 30 purified protein complexes. How accurate replication rates are achieved on a global scale and how chromatin factors interact with components of the replication machinery (right panel), remains poorly understood.
Selected publications
• Kurat CF, Yeeles JTP, Patel H, Early A and Diffley JFX (2017). Chromatin controls DNA replication origin selection, lagging strand synthesis and replication fork rates; Molecular Cell, 65(1):117-130
• Kurat CF, Lambert JP, Petschnigg J, Friesen H, Pawson, T, Rosebrock A, Gingras AC, Fillingham J and Andrews B (2014) Cell cycle regulated oscillator coordinates core histone transcription through histone acetylation; PNAS 111(39): 14124-9
• Kurat CF, Recht J, Radovani E, Durbic T, Andrews B and Fillingham J (2014) Regulation of histone gene transcription in yeast; Cell Mol Life Sci 71(4): 599-613
• Kurat CF, Lambert JP, van Dyk D, Tsui K, van Bakel H, Kaluarachchi S, Friesen H, Kainth P, Nislow C, Figeys D, Fillingham J and Andrews B (2011) Restriction of histone gene transcription to S phase by phosphorylation of a chromatin boundary protein; Genes & Development 25(23): 2489-501

