Regnard Group
Our aim is to understand how the coding part genome, that represents only a minute fraction of the whole genome in higher eucaryotes, is regulated and maintained at the level of chromatin.
Active genes are epigenetically encoded in each cell of a multicellular organism. The best characterized histone mark specifically enriched at active genes is a methylation of Lysine 36 on the histone H3 tail (H3K36me3). It is set (or “written”) co-transcriptionally by the histone methyltransferase Set2, while traveling along with elongating RNA polymerase II. Chromatin proteins that can specifically bind (or “read”) this epigenetic mark usually contain a so-called PWWP domain. This domain has an aromatic pocket containing two tryptophanes (WW), which allows binding to methylated Lysine 36 on the H3 tail.
The number of proteins with a PWWP domain vary greatly between species (Table 1). In flies, the nine PWWP domain-containing proteins are poorly described. Nonetheless, most of them have orthologous counterparts in humans.
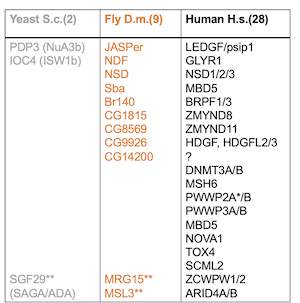
Table 1: Overview of potential/known H3K36me3 reader proteins harboring a PWWP domain in yeast, fly and human. The number of proteins is shown in brackets. For the two yeast proteins, the complexes they are associated with are mentioned in brackets. Among the nine PWWP domain-containing proteins in flies, four are only computed genes (GC) of unknown function. For human PWWP domain containing proteins, not all genes are listed and many of them have several paralogs.
*The PWWP domain of PWWP2A is required for binding H2A.Z containing nucleosomes.
**Proteins binding H3K36me3 with a chromodomain.
The function of H3K36me3 is not clearly elucidated. However, the replacement of the replication dependent histone H3.2 variant by a K36R mutant, which cannot be methylated be Set2 in flies, has shown that this residue of histone H3 is essential for viability after pupal stages, but only has mild defects in transcription. We would like to understand the role of epigenetically encoding gene activity, focusing on H3K36 methylation and the “readers” recruited to chromatin by this modification.
Signaling within the active genome
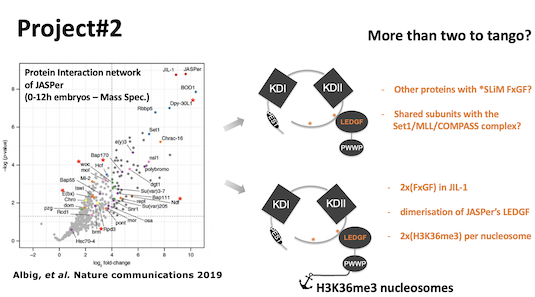
We recently characterized a chromatin-associated kinase complex in flies, which is tethered to the gene body of active genes. This JJ-kinase complex is composed of JASPer, orthologous to LEDGF/psip1 in humans, which stabilizes the essential kinase JIL-1, orthologous to the human Mitogen and Stress activated Kinases MSK-1/2 (Figure 1).
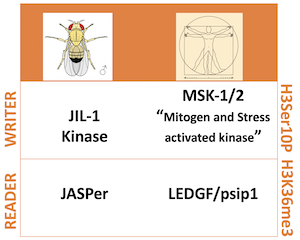
Figure 1: Orthologous interphase histone H3Ser10 phosphorylation “Writers” and H3K36me3 “Readers” in fly and human. No link is known yet between MSK-1/2 and LEDGF/psip1.
JASPer is stabilizing JIL-1 by binding to its conserved C-terminal domain with its LEDGF domain, and the PWWP domain is responsible for tethering the kinase complex to nucleosomes with H3K36me3. Thus JASPer controls the distribution and abundance of JIL-1 dependent phosphorylation, in particular the interphase specific histone H3 phosphorylation. However, in vitro assays have shown that H3K36me3 is not the signal that leads to activation of the kinase complex in the chromatin context (Albig, et al. Nature communications 2019).
In project 1, we are exploring which types of (stress-)signals lead to activation of JIL-1 kinase and which other chromatin components are JIL-1 substrates, in particular within the protein interaction network of JASPer and JIL-1 (see also project#2).
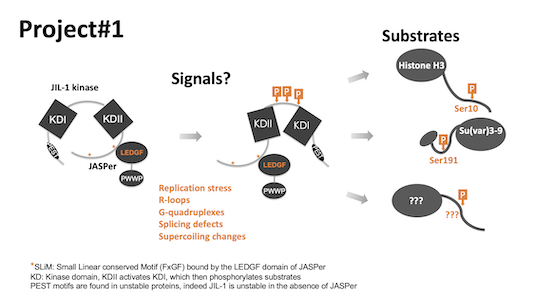
In project#2, we aim at reconstituting a functional kinase complex in vitro to study its activity on different types of substrates and in particular on various chromatin templates. We will employ a range of methods from reconstitution using only recombinant components, to more complex chromatin, assembled using early embryo extracts.