Projects
Histone Variants and Posttranslational Histone Modifications
Chromatin is formed by repetitive units called nucleosomes, which consist of ~ 146 base pairs of DNA wrapped around an octameric protein complex containing two copies of histones H2A, H2B, H3 and H4.
Many biological processes, such as gene regulation, mitosis, DNA repair and apoptosis are regulated by alterations of chromatin structure. Several different mechanisms exist to change the structure of chromatin, thereby influencing the functional output. These processes include methylation of the DNA itself, post-translational histone modifications (PTMs), remodeling of nucleosomes and the incorporation of specialized histone variants into nucleosomes (reviewed in Bonisch C. et al., 2008).
Posttranslational histone modifications are diverse and include lysine and arginine methylation; lysine acetylation; serine, threonine and tyrosine phosphorylation, ubiquitilation, SUMOylation, ADP-ribosylation and citrullination of methyl-arginine residues. Different theories have been developed to explain how these chemical marks might be translated into biological responses. The “histone code” hypothesis states that a specific histone modification, or combinations thereof, can affect distinct downstream cellular events by altering the structure of chromatin (cis mechanism) or by generating a binding platform for effector proteins (trans mechanism) (Strahl and Allis, 2000).
Furthermore, several histone variants that differ in amino acid sequence to their canonical counterparts, have evolved. These variants provide another mean of chromatin alterations, either by directly affecting the structure of the nucleosome or by introducing new modification sites (reviewed in Bernstein, E. and Hake, S.B. 2006)
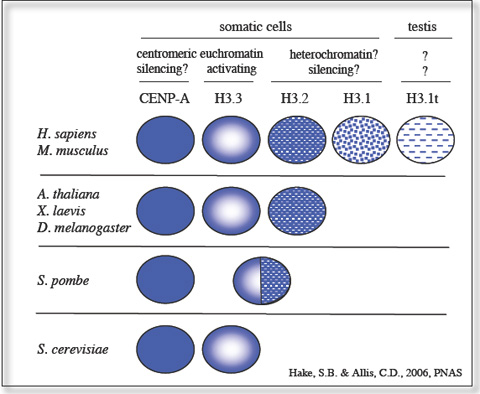
We are interested in understanding the biological functions of mammalian histone H3 variants and the role of different H3 post-translational modifications. Our goal is to identify the role of H3.1, H3.2 and H3.3 in different biological processes, in particular cell differentiation and gene regulation (Hake, S.B. and Allis, C.D. 2006) and to characterize novel H3 variants, which we termed H3.X and H3.Y, with the help of diverse biochemical approaches and bio-imaging techniques. Additionally, we are investigating the impact of several novel histone H3 PTMs on mitotic processes and gene regulation. Finally, we are using a variety of biochemical methods to identify potential PTM binding proteins to learn more about downstream effector cascades.
References:
Bernstein, E. and Hake, S.B. 2006. The nucleosome: a little variation goes a long way. 84(4): 505-517
Bonisch, C., Nieratschker, S.M., Orfanos N.K., and Hake, S.B. 2008. Chromatin proteomics and epigenetic regulatory circuits. Expert Rev. Proteomics, 5(1): 105-119
Hake, S.b. and Allis, C.D. 2006. Histone H3 variants and their potential role in indexing mammalian genomes: the “H3 barcode hypothesis”. 103(17): 6428-6435
Strahl, B.D. and Allis, C.D. 2000. The language of covalent histone modifications. Nature, 403(6765): 41-45
Projects:
1. Histone H3 Variants and their Functions
Mammals express at least five different histone H3 variants: H3.1, H3.2, H3.3, CENP-A, and H3.1t. What is the functional difference between the somatic H3.1, H3.2 and H3.3 variants and what roles do they play in gene regulation?
2. Novel H3 Variants: H3.X and H3.Y
Recently, we identified two novel putative H3 variants, which we termed H3.X and H3.Y. Where and when are these variants expressed and what is their function?
3. Posttranslational histone modifications during mitosis
During mitosis histones are increasingly phosphorylated on several different residues. What is role of H3.3S31ph during metaphase and are there any novel modifications found during mitosis?
4. Novel posttranslational modifications on histone H3
During previous MS/MS studies we have identified novel, uncharacterized posttranslational modifications on histone H3. What are the "writers", "readers", "erasers" and functions of these marks?

