Research Interests
We explore the principles and mechanisms of dynamic regulation of genome functions through chromatin organisation. Our model organism is Drosophila melanogaster. We employ a wide range of methods from biochemistry, genomics, cell biology, reverse genetics and collaborate with colleagues on structural aspects. One speciality is the in vitro reconstitution of various types of chromatin.
Targeting the X chromosome for global transcription activation
Transcription factors are recruited to their target sites by DNA sequence. However, genomes commonly contain many sequences that look like binding sites, but only a fraction of them are truly functional. The phenomenon of X chromosome dosage compensation in Drosophila provides a unique opportunity to explore targeting principles. The key regulator, the male-specific-lethal dosage compensation complex (MSL-DCC), selectively binds to ‘High Affinity Sites’ on the X chromosome in male cells. It then doubles the transcription output of most genes on this chromosome to match the expression of the two Xs in female cells. We explore how DNA structure, chromatin and cooperativity between various DNA-binding principles leads to binding sites selection. A central methodological approach is to reconstitute the X-autosome discrimination in cell-free systems.
further reading:
Prayitno K, Schauer T, Regnard C & Becker PB (2019). Progressive dosage compensation during Drosophila embryogenesis is reflected by gene arrangement. EMBO Reports 8, e48138.
Albig C, Tikhonova E, Krause S, Maksimenko O, Regnard C, Becker PB (2019). Factor cooperation for chromosome discrimination in Drosophila. Nucleic Acids Research 47, 1706-1724.
Schauer T, Ghavi-Helm Y, Sexton T, Albig C, Regnard C, Cavalli G, Furlong EE & Becker PB (2017). Chromosome topology guides the Drosophila Dosage Compensation Complex for target gene activation. EMBO Reports pii: e201744292.
Villa R, Schauer T, Smialowski P, Straub T & Becker PB (2016). PionX sites mark the X chromosome for dosage compensation. Nature 537, 244-248.
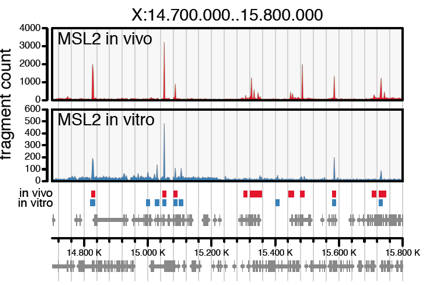
Figure taken from Villa et al., 2016.
The role of long, non-coding RNA in the dosage compensation complex
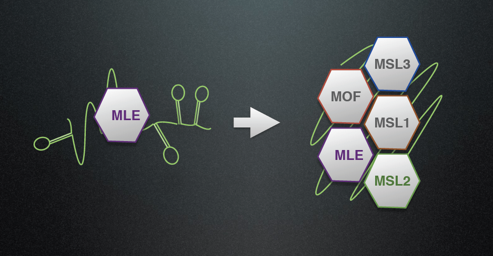
The MSL-DCC a complex made of five proteins (three enzymes) and long, non-coding RNA (roX RNA). We study the interactions of all recombinant components, their enzymatic activities and chromatin targeting specificity. We have shown that the RNA helicase MLE remodels repressive stem-loops in roX to expose binding sites for MSL subunits. We hypothesize that the assembling the DCC subunits around a dynamic RNA scaffold follows a distinct pathway that leads to a functional DCC. Our goal is to solve the pathway of DCC assembly and the influence roX aptamer have on the properties of associated proteins.
Further reading:
Müller M, Schauer T, Krause S, Villa R, Thomae AW, Becker PB (2020). Two-step mechanism for selective incorporation of lncRNA into a chromatin modifier.
Nucleic Acids Res 48:7483-7501.
Prabu JR*, Müller M*, Thomae AW, Schüssler S, Bonneau F, Becker PB* & Conti E* (2015). Structure of the RNA Helicase MLE Reveals the Molecular Mechanisms for Uridine Specificity and RNA-ATP Coupling. Molecular Cell 60, 487- 499.
Militti C, Maenner S, Becker PB & Gebauer F (2014). UNR facilitates the interaction of MLE with the lncRNA roX2 during Drosophila dosage compensation. Nature Commun. 5,4762-4769.
Maenner S, Müller M, Fröhlich J, Langer D & Becker PB (2013). ATP-dependent roX RNA remodeling by the helicase maleless enables specific association of MSL proteins. Molecular Cell 51, 174-184.
The chromatin fiber
Since our discovery of the Chromatin Accessibility Complex (CHRAC) and nucleosome sliding as a fundamental principle underlying chromatin dynamics we learnt a lot about the mechanism of nucleosome sliding. It is less clear what the physiological functions of these remodelers might be. We recently characterised cells and flies that lack functional ACF1, the signature subunit of ACF and CHRAC. Currently, we focus on the role of CHRAC/ACF in nucleosome remodelling at sites of DNA damage.
Further reading:
Baldi S, Korber P, Becker PB (2020). Beads on a string-nucleosome array arrangements and folding of the chromatin fiber. Nat Struct Mol Biol. 27:109-118.
Harpprecht L, Baldi S, Schauer T, Schmidt A, Bange T, Robles MS, Kremmer E, Imhof A & Becker PB (2019). A Drosophila cell-free system that senses DNA breaks and triggers phosphorylation signalling. Nucleic Acids Research May 31, doi: 10.1093/nar/gkz473.
Baldi S, Jain DS, Harpprecht L, Zabel A, Scheibe M, Butter F, Straub T & Becker PB (2018). Genome-wide Rules of Nucleosome Phasing in Drosophila. Molecular Cell 72, 661-672.
Baldi S, Krebs S, Blum H & Becker PB (2018). Genome-wide measurement of local nucleosome array regularity and spacing by nanopore sequencing. Nature Struct Mol Biol. 25, 894-901
Scacchetti A, Brueckner L, Jain D, Schauer T, Zhang X, Schnorrer F, van Steensel B, Straub T & Becker PB (2018). CHRAC/ACF contribute to the repressive ground state of chromatin. Life Science Alliance 1, e201800024.
Domino complexes and the histone variant H2A.V
In Drosophila melanogaster the properties of the two ancient, ubiquitous histone H2A variants, H2A.X and H2A.Z, are combined in a single molecule, H2A.V. This variant functions as DNA damage sensor and as an architectural element of active promoters and it has further roles in heterochromatin formation. We wonder how a single histone variant can carry out such diverse functions. Key to answering this question is an understanding of the ATP-dependent nucleosome remodelling machines that are involved in placement and turnover of H2A.V. H2A variant exchange is catalysed by evolutionarily conserved SWR1-like remodeling enzymes, of which Drosophila has only one, the ATPase Domino. We found that two alternative DOM splice variants (DOM-A and DOM-B) that differ in their C-terminus have distinct cell type-specific functions in development and H2A.V exchange. Our aim is to characterise the subunit composition, chromatin binding and substrate specificity of DOM-A and DOM-B complexes.
Further reading:
Scacchetti A, Becker PB (2020). Variation on a theme: Evolutionary strategies for H2A.Z exchange by SWR1-type remodelers..Curr Opin Cell Biol.70:1-9.
Scacchetti A, Schauer T, Reim A, Apostolou Z, Campos Sparr A, Krause S, Heun P, Wierer M, Becker PB (2020). Drosophila SWR1 and NuA4 complexes are defined by DOMINO isoforms.
Elife 9:e56325.
Börner K & Becker PB (2016). Splice variants of the SWR1-type nucleosome remodeling factor Domino have distinct functions during Drosophila melanogaster oogenesis. Development 143, 3154-67.
Baldi S & Becker PB (2013). The variant histone H2A.V of Drosophila-three roles, two guises. Chromosoma 122, 245-258

Figure taken from Börner et al. (2016)

